The fascinating world of proteins, with their intricate structures and functions, has attracted the attention of scientists for decades, and continues to do so more than ever.
Proteomics, the large-scale study of protein structure and function, has emerged as a powerful tool in this venture. Going one step further, quantitative proteomics, which identifies and quantifies proteins, offers a more detailed and dynamic view of the biological processes happening inside cells.
In this article, we provide an overview of quantitative-proteomics, its applications in biomedical and life sciences, and how we at Silantes are helping advance this field of research.
What is Proteomics?
Proteomics, derived from PROTEin complement of a genOME, is the large-scale study of protein structure and function.
The proteome – unlike the relatively static genome – is dynamic, changing in response to a myriad of factors like the environment, time, and stressors. Proteomics aims to comprehensively understand the complex protein interactions and changes that occur within cells, tissues, or organisms.
This makes it an essential tool across a broad spectrum of applications in the field of biomedical and life sciences.
In the area of biomarker discovery, quantifying the changes in protein abundance under different physiological and pathological conditions, aids in identifying specific markers that signal disease onset or progression.
They are also of great value in therapeutic target identification. By elucidating the complex network of protein interactions and modifications, quantitative proteomics enables researchers to identify key proteins involved in disease pathways.
In the field of drug discovery, insights into the effects of potential drugs on protein expression levels, help researchers assess the efficacy and possible side effects of novel therapeutics. This approach can streamline the drug discovery process, saving time and resources by allowing early identification of promising drug candidates and elimination of ineffective ones.
In order for scientists to elucidate information such as disease pathways or discover potential therapeutic targets, proteins must first be isolated, before various techniques are used to identify and quantify them.
While identifying the proteins present in a biological sample is crucial, it is only part of the story. For a comprehensive understanding, it is equally important to quantify these proteins.
Also, they are allowing the measurement of protein amounts over time and between different conditions and can indicate which proteins are upregulated or downregulated under certain conditions. This provides a more nuanced view of the proteome, going beyond just the presence or absence of proteins, to reveal a dynamic picture of biological activity.
The Role of Mass Spectrometry
Mass spectrometry (MS) has become an indispensable tool in quantitative proteomics. It is used to separate, identify, and quantify proteins based on their mass-to-charge ratio. The information it provides, coupled with bioinformatic data analysis, allows for a comprehensive picture of the proteome.
The key steps in a typical MS-based proteomics experiment include protein extraction, digestion into peptides, separation by liquid chromatography, and detection by mass spectrometry. Through a process known as tandem mass spectrometry (MS/MS), specific peptides can be selected for fragmentation, providing sequence information and enabling protein identification.
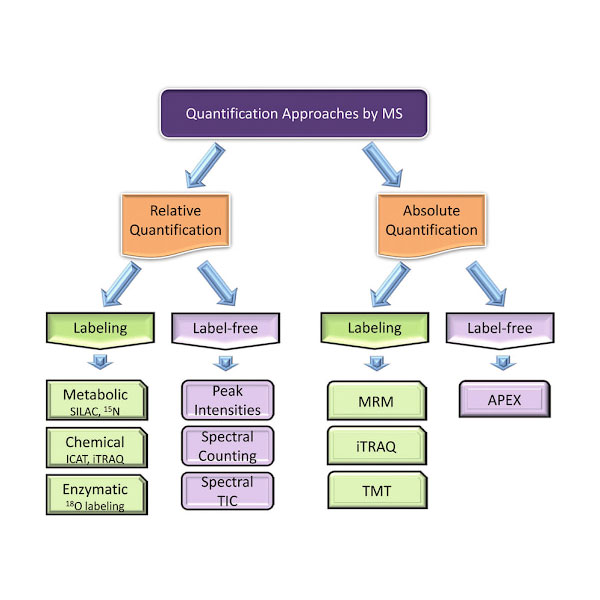
Relative & Absolute Quantification
In order to compare protein expression levels between different samples, both relative and absolute quantification methodologies are used.
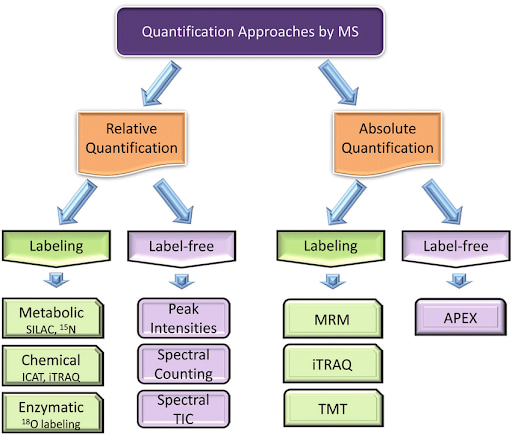
Relative quantification is typically based on differential labeling of proteins from two or more samples, allowing for comparison of their abundances. This approach allows for peptide signal intensities to be compared, hence determining peptide abundance across relative samples.
Absolute quantification on the other hand, allows for a precise measure of the quantity of a specific protein in a sample. For this purpose, known amounts of peptides are spiked into the sample.
Choosing how you wish to quantify proteins will depend on whether you use a label, or label-free methodology.
Label-Free Quantification
In label-free quantification, protein abundance is measured directly from the raw MS data without the use of any stable isotope labeling. Two common methods are spectral counting, where the number of identified spectra for a given protein is used as a proxy for abundance, and MS1 intensity-based methods, where the intensity of precursor ions is used to estimate protein quantities.
Isotope Labeling Quantification
Isotope labeling involves incorporating stable isotopes into proteins or peptides. This labeling creates a mass standard that can be used to compare to unlabeled proteins or peptides. Some of the commonly used labeling methods are outlined below:
Stable Isotope Labeling by Amino acids in Cell culture (SILAC)
In SILAC, cells are grown in a medium containing either ‘light’ or ‘heavy’ versions of specific amino acids. The resulting proteins are then combined and analyzed by MS, allowing for direct comparison of protein abundance between conditions.
Isotope-Coded Affinity Tag (ICAT)
ICAT labels cysteine residues on proteins with isotopically ‘light’ or ‘heavy’ tags. After labeling, the protein samples are mixed, digested, and the labeled peptides are selectively isolated and analyzed.
Tandem Mass Tags (TMT)
TMT is a method where peptide samples are labeled with different isobaric tags. After fragmentation in the mass spectrometer, each tag generates a unique reporter ion, which is used to quantify the peptides.
Isobaric Tags for Relative and Absolute Quantification (iTRAQ)
Similar to TMT, iTRAQ involves labeling peptides with isobaric tags. However, iTRAQ allows for the comparison of up to eight different samples in a single experiment, making it especially useful for larger-scale studies.
The Advantages
Quantitative proteomics provides a wealth of benefits for biological and medical research. First and foremost, it offers comprehensive profiling, delivering an in-depth overview of protein expression, post-translational modifications, and protein-protein interactions. This in turn facilitates a more profound understanding of biological processes and disease mechanisms.
Furthermore, it’s a powerful tool in the realm of biomarker discovery for diagnostics and therapeutics. By comparing healthy and diseased tissues, researchers can identify proteins that significantly change in abundance, pinpointing potential disease markers. In drug discovery, quantitative proteomics plays a pivotal role, helping to identify target proteins and evaluate the effect of drugs on protein expression levels.
Finally, given the modern mass spectrometry technologies, they can analyze thousands of proteins in a single experiment. This high-throughput analysis capability makes it an invaluable resource for large-scale proteome studies.
However, just like any scientific method, they have their limitations. Certain types of proteins are particularly difficult to analyze using current techniques. In addition, reproducing accurate and repeatable results, combined with interpretation of vast amounts of data is also another significant challenge facing scientists carrying out quantitative proteomics.
The Advantages of SILAC
SILAC is a powerful technique that has several advantages over other protein quantification methods.
SILAC’s in vivo labeling approach minimizes sample handling and avoids potential errors introduced during protein extraction, as the isotopic labeling occurs inside the cell before cell lysis. This ensures that every peptide of the protein of interest is labeled, providing higher accuracy.
Labeling cell cultures allows for labeling of all peptides in a sample, allowing for the quantification of the entire proteome and not just specific proteins. When analyzed via LC-MS using internal standards, it is possible to achieve a global view of protein expression changes under different conditions.
The ability to perform multiplex analysis reduces experimental variation and increases the throughput of data acquisition.
Conclusion
Quantitative proteomics represents a significant step forward in the understanding of biological systems and disease processes.
By combining the identification of proteins with their quantification, researchers can delve into the dynamic changes of the proteome, leading to a richer understanding of cellular mechanics. This knowledge, coupled with the ability to analyze thousands of proteins in a single experiment, has the potential to significantly contribute to areas like biomarker discovery, therapeutic target identification, and drug discovery.
While they are facing challenges in terms of complexity of biological samples, reproducibility, and data interpretation, ongoing advancements in technologies and bioinformatics are continually improving its accuracy and utility.
FAQs
What is quantitative proteomics?
Quantitative proteomics is a branch of proteomics that involves the identification and quantification of proteins in a sample, often using techniques like mass spectrometry. It allows researchers to compare the abundance of proteins between different samples, such as cells in a healthy state versus cells under disease conditions.
How does quantitative proteomics work?
Quantitative proteomics involves the measurement of the abundance of proteins in a sample. Some techniques involve labeling proteins or peptides with stable isotopes and comparing the intensities of labeled peptide ions.
Why is it important?
Quantitative proteomics plays a crucial role in understanding biological processes, providing insights into how these changes impact cell function and contribute to disease. This information can contribute to the discovery of new biomarkers for disease diagnosis and prognosis, as well as potential targets for drug development.
Why is proteomics more important than genomics?
While both proteomics and genomics are crucial for understanding biological systems, proteomics offers some unique advantages. The genome of an organism is by and large static, but the proteome is dynamic and changes in response to various factors such as environmental conditions, developmental stages, and disease states. By studying the proteome, we can gain a more detailed view of the cell’s functional state.
What are the applications of quantitative proteomics?
Quantitative proteomics has numerous applications in biomedical research, such as understanding disease mechanisms, identifying potential therapeutic targets and biomarkers, and monitoring drug responses.
What are the methods of quantitative analysis of proteins?
Labeling methods include SILAC, iTRAQ, and TMT. These techniques label proteins or peptides with isotopes, allowing for accurate comparison and quantification by mass spectrometry. Label-free methods estimate protein abundance from the number of spectra identified per protein or the intensity of peptide ions in the mass spectra.